Introduction to SNA: Concepts and Data
Who am I
- I’m Kevin Reuning (ROY-ning).
- I’m an Assistant Professor in Political Science.
- Prior to grad school I had very little experience in coding.

Goals For this Bootcamp
- Understand basic SNA terminology
- Load a variety of SNA formats into R.
- Calculate some basic network and nodal statistics.
- Make network visualizations.
- Know where to look for more.
Where We Are Going
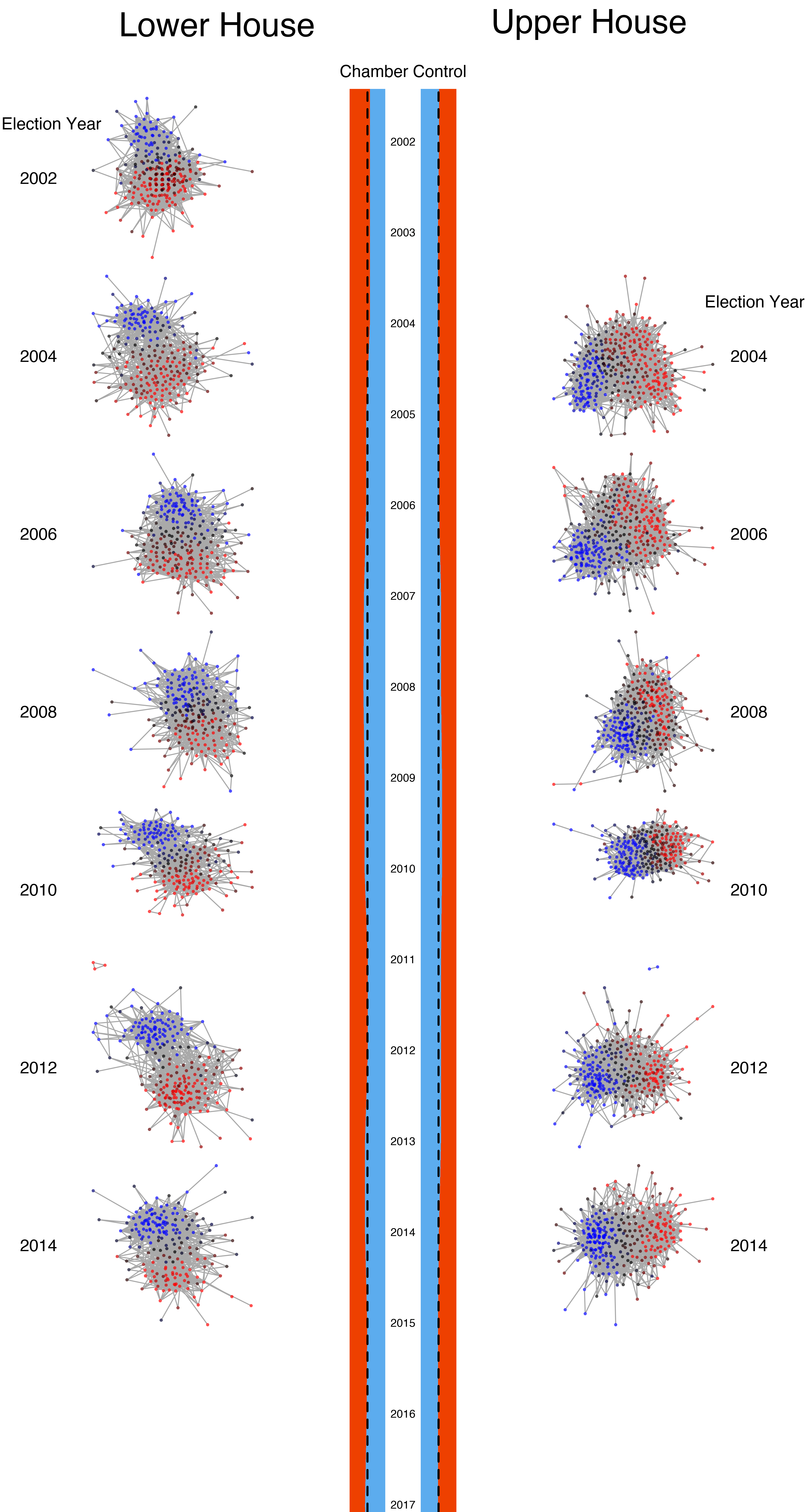
Goals for Today
- Go over some basic language of social network analysis.
- Load some SNA data into R.
- Start manipulating that
Nodes/Vertices and Edges
SNA focuses on the relationships between different entities:
- Nodes or Vertices: The entities that make-up your network.
- Ex: individuals, animals, organizations, counties, …
- Edges: The relationships that make-up your network.
- Ex: Friendship, proximity, exchange of goods, …
Edges - Variation and Types
Edges comes in many flavors and can be divided between states and events.
- States: The relationship is on-going (not forever necessarily but it exists overtime)
- Types: Similarities, Roles, Cognition
- Events: The relationship is captured by some discrete moment in time.
- Types: Interactions and Flows.
How Events relate to States
Often we use events to identify a state:
- Two students that are often seen together are likely to be friends.
- Two students that text often are likely to be friends.
We also can think that events lead to a state:
- Interacting with someone might lead to a friendship.
Edge Differences and Attributes
- Edges can be directed or undirected:
- Directed: Point from node A to node B.
- Undirected: Are between node A and node B.
- Edges can also be weighted. Examples:
- The amount of trade flowing from country A to country B.
- How long two individuals have known each other.
- The valence of a feelings towards another node (negative to positive)
Adjacency Matrix
Networks can be written out as adjacency matrix:
\[ \mathbf{A} = \left[\begin{array} {rrr} 0 & 0 & 1 & 0 \\ 0 & 0 & 0 & 0 \\ 1 & 1 & 0 & 1 \\ 1 & 0 & 1 & 0 \\ \end{array}\right] \]
Adjacency matrices are written in row-to-column format. Undirected networks will always have a symmetric matrix
Edge Lists
The other common way to make write out a network is through an edge list format:
| from | to |
|---|---|
| a | c |
| c | a |
| c | b |
| c | d |
| d | a |
| d | c |
R and SNA
There are two major sets of R libraries used for networking:
Following along
You need to learn by doing. If you haven’t opened RStudio yet, do so now.
Network Data Formats
There are a variety of ways that networks are saved/shared:
igraph Object
In each case we need to load in data and turn it into an igraph object.
As an igraph object we can easily apply a lot of network methods do it, plot it, etc.
Loading Network from an Adjacency Matrix
Start with a network of cocaine smugglers. Download the csv here, more info here
Need to do the following:
- Load the csv into R, treating the row names appropriately
read.csv()- Set
row.names=1so that the first column is read in as row names.
- Set
- Convert it to a matrix
as.matrix() - Convert it to an igraph object
graph_from_adjacency_matrix()
graph_from_adjacency_matrix()
There are some options we can set:
mode="directed"directed network"undirected"undirected, using upper triangle to make
weighted=NULL(default) the numbers in matrix give the number of edges between.TRUEcreates edge weights.NAcreates edges if they are greater than 0, ignore the rest.
diag=where to include the diagonal, set toFALSEto ignore diagonals.add.colnames=NULL(default) use column names as the vertex names.NAignore the column names.
igraph Object
Calling the igraph object by itself provides some details about the network, including some example edges:
IGRAPH ab1b6e0 DNW- 38 68 --
+ attr: name (v/c), weight (e/n)
+ edges from ab1b6e0 (vertex names):
[1] ABFM ->AFM ABFM ->FLMC ABFM ->JES ABFM ->JHY ABFM ->MCM ABFM ->RBM
[7] ABFM ->VFH AFM ->JES AIGC ->FFM AIGC ->JES CAR ->FFM DEJV ->CAR
[13] DMN ->ABFM FAERH->RBM FFM ->CAR FFM ->H5 FFM ->JES FFM ->M2
[19] FFM ->MRQ FFM ->RJJ FLMC ->ABFM FLMC ->DEJV FLMC ->EYVT FLMC ->H1
[25] FLMC ->H2 FLMC ->H3 FLMC ->H9 FLMC ->JAGG FLMC ->JES FLMC ->JFM
[31] FLMC ->ROB H10 ->FFM H11 ->ABFM H6 ->JES H7 ->JES H8 ->JES
[37] JAGG ->FLMC JES ->ABFM JES ->AFM JES ->AIGC JES ->CHA JES ->FFM
[43] JES ->FLMC JES ->JFM JES ->M3 JES ->RBM JFM ->ABFM JFM ->AFM
+ ... omitted several edgesBasic Functions
Basic Plot
We will make better plots later, but this gives us a quick idea of what our network looks like
Network Components
Dividing Up a Network
- We can group vertices by who they can reach:
- A component is the maximal (largest) group of vertices where every vertex within it can reach every other vertex.
- Every network can be broken into 1 or more component(s).
- A vertex that cannot reach any other vertices is an isolate
Some Components
This network has 6 components, the largest has 10 vertices in it.
Components with Directed Graphs
When a network is directed, then we need to think about direction.
- Weak Component: Is a component if we disregard direction of edges.
- Strong Component: Is a component if we follow direction of edges
Strong/Weak Components:
Calculating Components
[1] 22 [1] 1 1 1 1 1 1 1 1 1 17 1 1 1 1 1 1 1 1 1 1 1 1 ABFM AFM AIGC AMG CAR CHA DEJV DMN EYVT FAERH FFM FLMC H1
10 10 10 13 10 19 10 9 18 10 10 10 17
H10 H11 H2 H3 H5 H6 H7 H8 H9 JAGG JES JFM JHY
8 7 16 15 22 6 5 4 14 10 10 10 10
JMBM M1 M2 M3 MCM MRQ PR PRS RBM RJJ ROB VFH
12 3 21 10 10 10 2 1 10 20 10 11 Loading Network from an Edge List
Process
Now we are going to use a network of political donors in Ohio. Download the edgelist data here and the nodal data here. More info is here.
Need to do the following:
- Load the edge list and nodal data into R as two different objects
read.csv() - Combine them into an igraph object
graph_from_data_frame()
graph_from_data_frame
There are some options we can set:
directed=- Directed or not?
TRUEorFALSE
- Directed or not?
vertices=- Adding data to the vertices. The first column needs to match the identifiers used in the ede list.
Warning
You can only directly include isolates in edge lists if you have a vertex data frame.
igraph Object for Ohio Network
IGRAPH 6ae6587 UN-- 336 14183 --
+ attr: name (v/c), ContributorName (v/c), CatCodeIndustry (v/c),
| CatCodeGroup (v/c), CatCodeBusiness (v/c), PerDem (v/n), PerRep
| (v/n), DemCol (v/c), RepCol (v/c), Total (v/n), edge (e/c)
+ edges from 6ae6587 (vertex names):
[1] 10041 --1039 1025 --1039 10041 --1055 1025 --1055
[5] 1039 --1055 10041 --10680063 1055 --10688628 10680063--10701104
[9] 1025 --10770383 1039 --10770383 1025 --10812576 1039 --10812576
[13] 10770383--10812576 10041 --10986 1025 --10986 1039 --10986
[17] 1055 --10986 10680063--10986 10041 --1116 1039 --1116
[21] 1055 --1116 10680063--1116 10688628--1116 10986 --1116
+ ... omitted several edgesVertex and Edge Attributes
Accessing Them
You can access vertex and edges in your Igraph object using V() or E(). This is useful to access attributes using $variable
Example
There is a Total vertex attribute which is the total amount donated:
Deleting Vertices
This can be helpful in deleting vertices with the delete_vertices() function. Lets remove all vertices where they donated less than $2,000:
Deleting Edges
We can do the same thing with edges, lets keep just the edges that are marked "Strong" in the edge edge attribute:
Adding Attributes
We can also add an attribute to the network. Here we add vertex attribute that indicates what component everyone is in:
comps <- components(trimmed_net)
V(trimmed_net)$Comp <- LETTERS[comps$membership]
V(trimmed_net)$Comp[1:10] [1] "A" "A" "A" "A" "A" "A" "A" "A" "A" "A"comps$membership returns a numeric indicator of membership in a component. I use LETTERS[] to convert that into a letter instead of a number.
Other Formats
Loading Network Formatted Data
The read_graph() function can load in a variety of native network formats. You should set format= when you call it:
For example using this ground squirrel data
IGRAPH 3cc471b U-W- 60 340 --
+ attr: btw_soc (v/n), btw_spat (v/n), Node_All days_detected (v/n),
| stage_current_year (v/c), sex (v/c), fur_mark (v/c), id (v/c), weight
| (e/n)
+ edges from 3cc471b:
[1] 1-- 2 1-- 4 1-- 5 1--13 1-- 6 1-- 7 1--14 1--15 1-- 9 1--16 1--10 1--17
[13] 1--11 1--12 2-- 3 2-- 4 2-- 5 2-- 6 2-- 7 2-- 8 2-- 9 2--10 2--11 2--12
[25] 3--20 3--48 3-- 8 4--19 4--23 4-- 5 4-- 6 4-- 7 4--14 4--26 4--15 4-- 8
[37] 4-- 9 4--11 4--12 5--19 5--20 5--13 5-- 6 5-- 7 5--54 5--30 5--15 5-- 9
[49] 5--10 5--12 6--19 6-- 7 6--28 6--15 6-- 8 6-- 9 6--11 6--12 7--19 7--18
[61] 7--38 7--14 7--15 7-- 8 7-- 9 7--10 7--11 7--12 8--48 8-- 9 8--47 8--11
+ ... omitted several edgesPutting it Together
Test your Knowledge
The next slide has a bunch of datasets for networks. I want you to do the following:
- Find a network you think is interesting, download it.
- Open the network in R
- Calculate the number of vertices, edges, and the number of components.
- Create a vertex attribute for component membership.
Databases of Data
- Netzschleuder: https://networks.skewed.de/
- Click the “CSV” option to download them, the other formats use a strange file compression.
- This has some very large networks
- Animal Social Network Repository: https://bansallab.github.io/asnr/data.html
- Hosted on github, once you find the network, click on the “graphml” to show the data, and then there is a download button the right side with a downward arrow.
- UCINET Data: https://sites.google.com/site/ucinetsoftware/datasets
- Has canonical datasets, poorly maintained.
Social Network Concepts